Difference between revisions of "AMS Clinic5"
(→HR Topics) |
(→Lodging and Travel) |
||
| (36 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
* '''<span style="color:red">If you are interested in attending, please complete [https://docs.google.com/spreadsheet/embeddedform?formkey=dG5yZURQcjZXaGtZbzNvSDNmTWpON0E6MA this registration form]</span>''' | * '''<span style="color:red">If you are interested in attending, please complete [https://docs.google.com/spreadsheet/embeddedform?formkey=dG5yZURQcjZXaGtZbzNvSDNmTWpON0E6MA this registration form]</span>''' | ||
| + | |||
== Motivation and Format of the Clinic == | == Motivation and Format of the Clinic == | ||
| Line 10: | Line 11: | ||
The 5th AMS Instrument and Data Analysis Clinic and [https://sites.google.com/site/citofms/um/boulder-2013 2nd Aerodyne CIMS Meeting] and Clinic will take place in the Spring of 2013 at the University of Colorado-Boulder. They are organized by the [http://cires.colorado.edu/jimenez-group/ Jimenez Group] and [http://www.aerodyne.com Aerodyne], and also sponsored by [http://cires.colorado.edu CIRES]. | The 5th AMS Instrument and Data Analysis Clinic and [https://sites.google.com/site/citofms/um/boulder-2013 2nd Aerodyne CIMS Meeting] and Clinic will take place in the Spring of 2013 at the University of Colorado-Boulder. They are organized by the [http://cires.colorado.edu/jimenez-group/ Jimenez Group] and [http://www.aerodyne.com Aerodyne], and also sponsored by [http://cires.colorado.edu CIRES]. | ||
| + | |||
| + | [[File:Mass-defect.png |thumb| 200px | More data analysis deserts]] | ||
| + | [[File:IMG 0758.JPG |thumb |200px | Jose, Donna and Doug]] | ||
== Detailed Clinic Program == | == Detailed Clinic Program == | ||
| Line 29: | Line 33: | ||
*** 9am - 10am: High resolution | *** 9am - 10am: High resolution | ||
*** 10am - 5pm: PMF | *** 10am - 5pm: PMF | ||
| − | *** 5:30pm: happy hour at | + | *** 5:30pm: happy hour at [http://www.themedboulder.com/ The Med restaurant] (1002 Walnut St., Boulder) [https://maps.google.com/maps?q=the+med+boulder&fb=1&gl=us&hq=the+med&hnear=0x876b8d4e278dafd3:0xc8393b7ca01b8058,Boulder,+CO&cid=0,0,1407699258478646510&t=m&z=16&iwloc=A Map here]. |
| − | ** Fri Mar 29: post-meeting hiking or skiing for interested folks | + | ** Fri Mar 29: post-meeting hiking or skiing for interested folks, coordinated by Matt Coggon and Yu Jun Leon |
== Hardware & DAQ topics == | == Hardware & DAQ topics == | ||
| Line 89: | Line 93: | ||
* O:C ratio and CCN closure (Deepika Bhattu) | * O:C ratio and CCN closure (Deepika Bhattu) | ||
* Fitting of SP-AMS peaks in the mid m/z range (Ed Fortner) | * Fitting of SP-AMS peaks in the mid m/z range (Ed Fortner) | ||
| − | * | + | * HR detection limits (Pedro Campuzano-Jost) |
* HR Frags (Pedro Campuzano-Jost) | * HR Frags (Pedro Campuzano-Jost) | ||
* HR analysis (Celia Faiola) | * HR analysis (Celia Faiola) | ||
| − | ''' Recipe for a | + | |
| + | ''' Recipe for a mass defect plot (you can choose to put a chocolate cake underneath it or not :) )''' | ||
** We want at these 4 items for the "mise en place" | ** We want at these 4 items for the "mise en place" | ||
** (A) The x axis wave is the exact mass wave, the wave of all the ions that were chosen to fit. | ** (A) The x axis wave is the exact mass wave, the wave of all the ions that were chosen to fit. | ||
** (B)The y axis wave is the mass defect wave, the wave that indicates the difference between the a pure carbon fragment and the mass offsets due to the Hs, Cs, etc. | ** (B)The y axis wave is the mass defect wave, the wave that indicates the difference between the a pure carbon fragment and the mass offsets due to the Hs, Cs, etc. | ||
| − | ** (C)The f(z) wave for the size of the markers will be the sqrt(log(HR peak stick)) | + | ** (C)The f(z) wave for the size of the markers will be the sqrt(log(HR peak stick)). However, log of stick values may be negative, and the sqrt of a negative is a nans, so we will have to adjust this wave that reflects size (usually markers, hence the sqrt) to be non-nan. |
** (D)The f(z) wave for the color of the markers will be the family colors | ** (D)The f(z) wave for the color of the markers will be the family colors | ||
| − | The first set of instructions will be a mass defect plot to be made for the sticks calculated by the gold button press "Calc HR sticks for one set of spectra | + | The first set of instructions will be a mass defect plot to be made for the sticks calculated by the gold button press "Calc HR sticks for one set of spectra |
| − | |||
* duplicate/o root:HR_sticks:ExactMassWave root:OneRunMassDefect | * duplicate/o root:HR_sticks:ExactMassWave root:OneRunMassDefect | ||
* root:OneRunMassDefect = root:HR_sticks:ExactMassWave - round(root:HR_sticks:ExactMassWave) | * root:OneRunMassDefect = root:HR_sticks:ExactMassWave - round(root:HR_sticks:ExactMassWave) | ||
| − | + | * duplicate/o root:HR_sticks:OneRunStickDiff root:LogOneRunDiffStick | |
| − | * duplicate/o root:HR_sticks:OneRunStickDiff root: | + | * LogOneRunDiffStick = log(root:HR_sticks:OneRunStickDiff |
| − | * | ||
| − | |||
* duplicate/o root:HR_sticks:OneRunStickDiff OneRunFamilyColorCode | * duplicate/o root:HR_sticks:OneRunStickDiff OneRunFamilyColorCode | ||
* OneRunFamilyColorCode = sq_findStr(root:HR:HR_familyName,root:HR_sticks:ExactMassFamilyText[p],1) | * OneRunFamilyColorCode = sq_findStr(root:HR:HR_familyName,root:HR_sticks:ExactMassFamilyText[p],1) | ||
| + | |||
| + | |||
| + | // Now to make the graph & prettify it: | ||
| + | * display OneRunMassDefect vs root:HR_sticks:ExactMassWave | ||
| + | * ModifyGraph zmrkSize(OneRunMassDefect)={LogOneRunDiffStick,*,*,1,10} | ||
| + | * ModifyGraph zColor(OneRunMassDefect)={OneRunFamilyColorCode,*,*,dBZ21,0} | ||
| + | * Label left "mass defect"; | ||
| + | * Label bottom "m/z" | ||
| + | * ModifyGraph mode=3,marker=19 | ||
| + | The second set of instructions will be a mass defect plot to be made for the sticks calculated with an HR spectra of species (i.e. from the results tab... in this example my wave is called MSSM_all_HROrg) | ||
| + | * duplicate/o root:HR_frag:SpeciesMassWave root:MassDefect_MSSM_all_HROrg | ||
| + | * root:MassDefect_MSSM_all_HROrg = root:HR_frag:SpeciesMassWave - round(root:HR_frag:SpeciesMassWave) | ||
| + | * duplicate/o root:MSSM_all_HRorg root:Log_MSSM_all_HROrg | ||
| + | * Log_MSSM_all_HROrg = log(root:MSSM_all_HROrg) | ||
| + | * duplicate/o root:HR_sticks:OneRunStickDiff MSSM_all_HROrg_FamilyColorCode | ||
| + | * MSSM_all_HROrg_FamilyColorCode = sq_findStr(root:HR:HR_familyName,root:HR_frag:SpeciesFamilyText[p],1) | ||
| − | Now to make the graph | + | |
| − | * ModifyGraph zmrkSize( | + | // Now to make the graph & prettify it: |
| − | * ModifyGraph zColor( | + | * display MassDefect_MSSM_all_HROrg vs root:HR_frag:SpeciesMassWave |
| + | * ModifyGraph zmrkSize(MassDefect_MSSM_all_HROrg )={Log_MSSM_all_HROrg,*,*,1,10} | ||
| + | * ModifyGraph zColor(MassDefect_MSSM_all_HROrg)={MSSM_all_HROrg_FamilyColorCode,*,*,dBZ21,0} | ||
* Label left "mass defect"; | * Label left "mass defect"; | ||
* Label bottom "m/z" | * Label bottom "m/z" | ||
| + | * ModifyGraph mode=3,marker=19 | ||
| + | |||
| + | Serve warm and with friends - Enjoy!! | ||
== PMF Topics == | == PMF Topics == | ||
| Line 126: | Line 149: | ||
* "PMF data processing" (Jianzhong Xu) | * "PMF data processing" (Jianzhong Xu) | ||
* Resolution of small factors in PMF (Misha Schurman) | * Resolution of small factors in PMF (Misha Schurman) | ||
| + | * Presentations shown | ||
| + | ** [http://cires.colorado.edu/jimenez-group/UsrMtgs/clinic_2013/2013-03-28_ME2_AMS_clinic_.pdf Jay Slowik: ME-2 Igor Tool] | ||
== Details of 5th AMS Clinic == | == Details of 5th AMS Clinic == | ||
| Line 137: | Line 162: | ||
* Feedback from past Clinics has been very positive overall, and we try to learn every time how to make the Clinic more useful and interesting to the users. You you can see the details for [http://cires.colorado.edu/jimenez-group/UsrMtgs/HRclinic1/2009-06_HR_Clinic_Feedback.pdf 2009], [http://cires.colorado.edu/jimenez-group/UsrMtgs/2ndClinic/2010-03_2ndClinic_Survey_Results_Complete.pdf 2010], and [http://cires.colorado.edu/jimenez-group/UsrMtgs/clinic_2011/2011_Clinic_Feedback.pdf 2011], and [http://cires.colorado.edu/jimenez-group/UsrMtgs/4thClinic/2012_AMS_Clinic_Feedback.pdf 2012]. | * Feedback from past Clinics has been very positive overall, and we try to learn every time how to make the Clinic more useful and interesting to the users. You you can see the details for [http://cires.colorado.edu/jimenez-group/UsrMtgs/HRclinic1/2009-06_HR_Clinic_Feedback.pdf 2009], [http://cires.colorado.edu/jimenez-group/UsrMtgs/2ndClinic/2010-03_2ndClinic_Survey_Results_Complete.pdf 2010], and [http://cires.colorado.edu/jimenez-group/UsrMtgs/clinic_2011/2011_Clinic_Feedback.pdf 2011], and [http://cires.colorado.edu/jimenez-group/UsrMtgs/4thClinic/2012_AMS_Clinic_Feedback.pdf 2012]. | ||
| − | * Pictures from the | + | * Pictures from the 5th AMS Clinic can be found [https://picasaweb.google.com/103806884733971442452/AMSCIMSClinic2013?authuser=0&authkey=Gv1sRgCJSEzJnd79nVwwE&feat=directlink here]. |
== Logistics == | == Logistics == | ||
| Line 146: | Line 171: | ||
* The University of Colorado at Boulder will be on spring break the week of Monday March 25 - Friday March 29, so there will be minimal traffic. | * The University of Colorado at Boulder will be on spring break the week of Monday March 25 - Friday March 29, so there will be minimal traffic. | ||
| − | |||
* If you may be interested in attending the AMS Clinic, you should add yourself to the [http://groups.google.com/group/ams_clinic AMS Clinic Email List]. A shortcut to that page is [http://tinyurl.com/list-clinic http://tinyurl.com/list-clinic]. | * If you may be interested in attending the AMS Clinic, you should add yourself to the [http://groups.google.com/group/ams_clinic AMS Clinic Email List]. A shortcut to that page is [http://tinyurl.com/list-clinic http://tinyurl.com/list-clinic]. | ||
| Line 153: | Line 177: | ||
*General information on Boulder is [http://www.bouldercoloradousa.com/ here] | *General information on Boulder is [http://www.bouldercoloradousa.com/ here] | ||
| − | * You will most likely travel via [http://www.flydenver.com/ Denver International Airport] | + | ** You will most likely travel via [http://www.flydenver.com/ Denver International Airport] |
| − | * You can use the [https://www.supershuttle.com/default.aspx?GC=NCAR1 Supershuttle service] between the airport and the hotel and back (once in Boulder, you can walk to and from the meeting) | + | ** You can use the [https://www.supershuttle.com/default.aspx?GC=NCAR1 Supershuttle service] between the airport and the hotel and back (once in Boulder, you can walk to and from the meeting) |
| − | * | + | ** If you are looking for someone to share a room with we have created a spreadsheet for coordinating and posting contact information [http://tinyurl.com/Clinic-Roomshare here] |
| − | |||
| − | * If you are looking for someone to share a room with we have created a spreadsheet for coordinating and posting contact information [http://tinyurl.com/Clinic-Roomshare here] | ||
*[http://www.millenniumhotels.com/millenniumboulder/index.html Millenium] (recommended, closest to meeting location) | *[http://www.millenniumhotels.com/millenniumboulder/index.html Millenium] (recommended, closest to meeting location) | ||
Latest revision as of 17:31, 1 April 2015
A shorcut to this page is http://tinyurl.com/AMSClinic5
- If you are interested in attending, please complete this registration form
Contents
Motivation and Format of the Clinic
The Clinic is a HANDS-ON discussion-based workshop. The people who should attend the AMS Clinic are those who are directly using the AMS and doing (or are VERY involved in directly supervising) AMS data analysis of field or lab data. The clinic will be much less useful for PIs who are not directly involved in directly using the instrument and doing data analysis, or for people who are just starting to learn about the AMS but haven't done serious instrument use and data analysis yet. Attendees are expected to bring to the Clinic datasets in which they are currently working, and be ready to use them as a basis for discussion with the group.
Dates and Location
The 5th AMS Instrument and Data Analysis Clinic and 2nd Aerodyne CIMS Meeting and Clinic will take place in the Spring of 2013 at the University of Colorado-Boulder. They are organized by the Jimenez Group and Aerodyne, and also sponsored by CIRES.
Detailed Clinic Program
- The time allocation to different topics is based on the survey of attendees here.
- AMS Clinic: Tue Mar 25‐ Thu March 28
- Mon Mar 25
- 1-5 pm: 2nd Symposium on Advances in Atmospheric Mass Spectrometry
- 7 pm: Aerodyne Dinner @ Backcountry Pizza (2139 Arapahoe Ave, near the Millenium Hotel and CU, see Map
- Tue Mar 26
- 9am - 2:30: Hardware
- 2:30-5:30 pm: Quantification & UMR / Squirrel
- Wed Mar 27
- 9am - 11am: Quantification & UMR / Squirrel
- 11am-5:30 pm: High-resolution
- 6:30pm-11pmish: Party @ Donna's place in East Boulder, 875 Roxwood Lane #A, Boulder . Come any time after 6:30, food will arrive at 7pm. We will order pizza for which $ can be contributed; folks can bring their drink of choice. Informal music session by a few folks in the AMS community.
- Thu Mar 28:
- 9am - 10am: High resolution
- 10am - 5pm: PMF
- 5:30pm: happy hour at The Med restaurant (1002 Walnut St., Boulder) Map here.
- Fri Mar 29: post-meeting hiking or skiing for interested folks, coordinated by Matt Coggon and Yu Jun Leon
- Mon Mar 25
Hardware & DAQ topics
- DAQ
- Version 4 of DAQ (Joel Kimmel)
- Tutorial on how DAQ acquires data and saves it into HDF files (Matt Coggon)
- Variability over time of chopper positions (Eleanor Waxman)
- Hardware components
- Updates on instrument upgrades (chopper, vaporizer) (Amber Ortega)
- "Hardware issues" (ABHISHEK CHAKRABORTY)
- C-ToF issues (Matt Coggon)
- Ringing issue (Yu Jun Leong)
- Vaporizer temperature (Yu Jun Leong)
- Filament emission (Abishek & Veronique)
- Filament change (Celia Faiola)
- Shipping the AMS (Veronique)
- PToF issues (Veronique)
- LS signals (Jay Slowik)
- Presentations shown
- John Jayne on TOF Thresholds
- Joel Kimmel on Thresholds and other TOF concepts
- Filament replacement presentation
- Q-AMS Manual (has procedure for replacing filaments and servo)
Quantification & UMR Topics
- Frag table
- UMR frag tables for chamber experiments (Anthony Carrasquillo)
- Frag table adjustments (Amber Ortega)
- Gases other than air
- Quantification when using variable amounts of N2 and Ar in gas (Raea Hicks)
- Frag table when using gases other than air (Brett Palm)
- PToF
- How to deal with small negative diameters in PToF (Weiwei Hu)
- PToF trigger correction based on AB (Pedro Campuzano-Jost)
- PToF DC Marker & Air interference issues (Lu Xu)
- PToF DC marker issues (Deepika Bhattu)
- Other Topics
- RIE of SO4 (Phil Croteau)
- SMPS intercomparison (Brett Palm)
- How to dig into Squirrel code & make changes (Matt Coggon)
- High signals in m/z 120-127 in ACSM, from naphthalene (Weiwei Hu)
- Measurements of H2O content of aerosols (Shunsuke Nakao)
- Issues with false doublets in IE Cals (Pedro Campuzano Jost)
- Dealing with high loadings and/or large dynamic range in loadings (L Alexander)
- Chamber vs ambient timing in SOAS (Yu Jung)
- Presentations shown
HR Topics
- Donna Sueper: intro to new faster and better HR fitting
- HR fitting issues (ABHISHEK CHAKRABORTY)
- HR fitting (Pedro Campuzano-Jost)
- Calibration and fitting at high m/z (Liz Alexander)
- Assigment of unusual mass peaks from LA-AMS (L Alexander)
- Manjula Canagaratna: Update on O/C calibration and proposed new procedure
- O:C ratio and CCN closure (Deepika Bhattu)
- Fitting of SP-AMS peaks in the mid m/z range (Ed Fortner)
- HR detection limits (Pedro Campuzano-Jost)
- HR Frags (Pedro Campuzano-Jost)
- HR analysis (Celia Faiola)
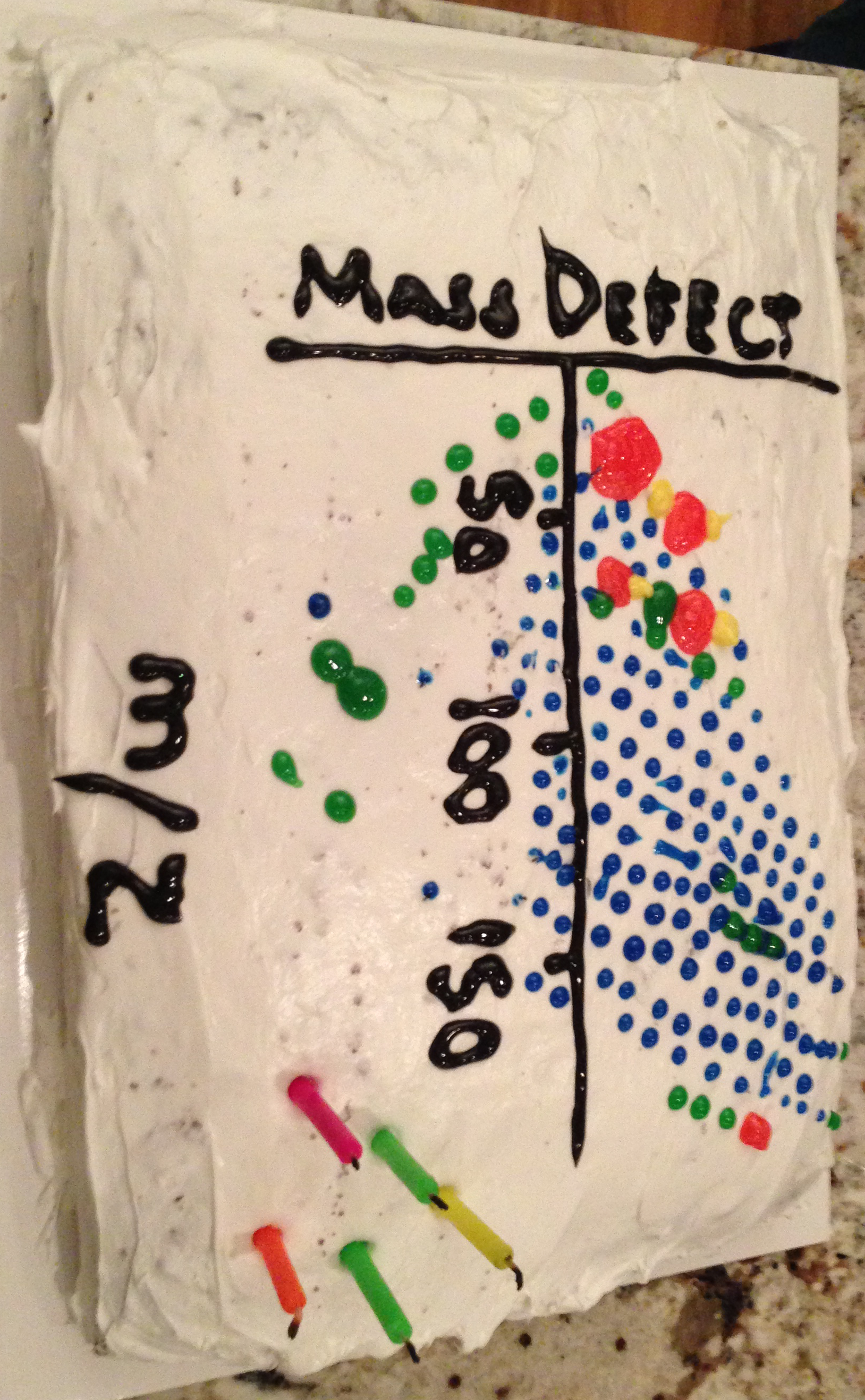
Recipe for a mass defect plot (you can choose to put a chocolate cake underneath it or not :) )
- We want at these 4 items for the "mise en place"
- (A) The x axis wave is the exact mass wave, the wave of all the ions that were chosen to fit.
- (B)The y axis wave is the mass defect wave, the wave that indicates the difference between the a pure carbon fragment and the mass offsets due to the Hs, Cs, etc.
- (C)The f(z) wave for the size of the markers will be the sqrt(log(HR peak stick)). However, log of stick values may be negative, and the sqrt of a negative is a nans, so we will have to adjust this wave that reflects size (usually markers, hence the sqrt) to be non-nan.
- (D)The f(z) wave for the color of the markers will be the family colors
The first set of instructions will be a mass defect plot to be made for the sticks calculated by the gold button press "Calc HR sticks for one set of spectra
- duplicate/o root:HR_sticks:ExactMassWave root:OneRunMassDefect
- root:OneRunMassDefect = root:HR_sticks:ExactMassWave - round(root:HR_sticks:ExactMassWave)
- duplicate/o root:HR_sticks:OneRunStickDiff root:LogOneRunDiffStick
- LogOneRunDiffStick = log(root:HR_sticks:OneRunStickDiff
- duplicate/o root:HR_sticks:OneRunStickDiff OneRunFamilyColorCode
- OneRunFamilyColorCode = sq_findStr(root:HR:HR_familyName,root:HR_sticks:ExactMassFamilyText[p],1)
// Now to make the graph & prettify it:
- display OneRunMassDefect vs root:HR_sticks:ExactMassWave
- ModifyGraph zmrkSize(OneRunMassDefect)={LogOneRunDiffStick,*,*,1,10}
- ModifyGraph zColor(OneRunMassDefect)={OneRunFamilyColorCode,*,*,dBZ21,0}
- Label left "mass defect";
- Label bottom "m/z"
- ModifyGraph mode=3,marker=19
The second set of instructions will be a mass defect plot to be made for the sticks calculated with an HR spectra of species (i.e. from the results tab... in this example my wave is called MSSM_all_HROrg)
- duplicate/o root:HR_frag:SpeciesMassWave root:MassDefect_MSSM_all_HROrg
- root:MassDefect_MSSM_all_HROrg = root:HR_frag:SpeciesMassWave - round(root:HR_frag:SpeciesMassWave)
- duplicate/o root:MSSM_all_HRorg root:Log_MSSM_all_HROrg
- Log_MSSM_all_HROrg = log(root:MSSM_all_HROrg)
- duplicate/o root:HR_sticks:OneRunStickDiff MSSM_all_HROrg_FamilyColorCode
- MSSM_all_HROrg_FamilyColorCode = sq_findStr(root:HR:HR_familyName,root:HR_frag:SpeciesFamilyText[p],1)
// Now to make the graph & prettify it:
- display MassDefect_MSSM_all_HROrg vs root:HR_frag:SpeciesMassWave
- ModifyGraph zmrkSize(MassDefect_MSSM_all_HROrg )={Log_MSSM_all_HROrg,*,*,1,10}
- ModifyGraph zColor(MassDefect_MSSM_all_HROrg)={MSSM_all_HROrg_FamilyColorCode,*,*,dBZ21,0}
- Label left "mass defect";
- Label bottom "m/z"
- ModifyGraph mode=3,marker=19
Serve warm and with friends - Enjoy!!
PMF Topics
- Ingrid on upgrades to the PET (error calculation, new plots)
- New PSI ME-2 Igor tool (Jay Slowik)
- PMF of flux data (Ben Langford)
- "PMF data processing" (Jianzhong Xu)
- Resolution of small factors in PMF (Misha Schurman)
- Presentations shown
Details of 5th AMS Clinic
- As planned, this is "out of phase" with the AMS Users Meeting and the heavy Fall meeting season (AAAR, EAC, AMS Users Meeting, AGU...).
- There is no registration fee, but please register if you are coming to plan food, coffee, tables, power, internet, name tags, hotel room block, etc.
- Information from previous AMS clinics (including presentations) are linked here: 1st AMS Clinic, 2nd AMS Clinic, 3rd AMS Clinic, and 4th AMS Clinic.
- Feedback from past Clinics has been very positive overall, and we try to learn every time how to make the Clinic more useful and interesting to the users. You you can see the details for 2009, 2010, and 2011, and 2012.
- Pictures from the 5th AMS Clinic can be found here.
Logistics
- The location will be the Ekeley Building in the CU Campus in Boulder. The location is linked here in Google Maps and here in the CU online map. The Northeast door of the building will be the only door open on Sunday. The room is Ekeley E1B20, just to the left after entering the building. It is the same location as last year.
- All AMS days will start at 9 am and go till 5-6 pm, with a break for lunch and additional coffee breaks.
- The University of Colorado at Boulder will be on spring break the week of Monday March 25 - Friday March 29, so there will be minimal traffic.
- If you may be interested in attending the AMS Clinic, you should add yourself to the AMS Clinic Email List. A shortcut to that page is http://tinyurl.com/list-clinic.
Lodging and Travel
- General information on Boulder is here
- You will most likely travel via Denver International Airport
- You can use the Supershuttle service between the airport and the hotel and back (once in Boulder, you can walk to and from the meeting)
- If you are looking for someone to share a room with we have created a spreadsheet for coordinating and posting contact information here
- Millenium (recommended, closest to meeting location)
- Rate: $109.00 for one King / $109.00 for two doubles (we are currently holding 10 rooms)
- Reservations: register online
- Contact p.303.443.3850 / CU - CIRES (for discounted rate)
- Reservations can only be made up to March 20, 2013 (for guaranteed rate)
- Boulder Inn (farther from meeting location, but a little cheaper)
- Rate: $89.00 for one King bed / $99.00 for two Queen Beds
- Contact: p.800.233.8469 / AMS / CIMS (for discounted rate)
- Reservations can only be made up to February 22, 2013 (for guaranteed rate)
- Boulder Outlook
- Rate: $69.00 for standard outside / $79.00 inside by pool / $89.00 executive (can either be a king or two doubles)
- Contact: p.800.542.0304 ext. 0 / ask for 'CU / CIRES AMS group' rate (must call hotel to get rate, cannot reserve online)
- Quality Inn
- Rate: $110.00 one queen / $118.00 one King / $124.00 two queens
- Contact: 1.888.449.7550 / ask for AMS / CIMS Clinic Rates
- If you want to find a roommate for the Clinic, please enter your information in this spreadsheet.
Please feel free to contact Michael Lechner (michael.lechner(at)colorado.edu for any further question)