Pk nextGen
Contents
Discussion: Next-generation PIKA
What we know for sure:
1. MikeC is to develop a PIKA-esque tool for multi-dimensional data from TofDAQ data files.
2. The non-AMS data format and multi-dimensional aware criteria forces a re-write of the HR code rather than straight adoption of existing PIKA
3. Other TW (non-AMS) applications will wish to use many existing PIKA features (not limited to PWPS, HRfrag, families, isotopic constraints), but others will be less applicable (obviously, the AMS-centric parts!).
4. Whatever gets written should address existing desires and concerns with the PIKA code, and be fully transparent so as to accept any input data type. It should be developed with memory and speed optimisation in mind.
What Mike initially proposed:
- An ion indexing scheme to allow saving/comparison of different ion fits at a given unit mass
- A dimension in the HR data set to save multiple copies of HR fits (this could be for different calibration, PW, PS, or simply open/closed AMS data)
- Fitting of subsets of unit masses within the mass spectrum (but over any dimensionality in time) to speed the process.
Concerns and discussion
Isotopic constraining
Summary of discussion thus far:
- Isotopic contraints, in practice, force the calculation of ion fits for every unit mass in the spectrum to be performed together, and in the correct order
- The fitting of a single unit mass at a time, whilst potentially useful in speed, is not thus not practical
The system is thus
- constrained to calculate all the fits together, meaning
- speed optimisation must come from assessing current memory management and use of threadsafe functions
- a lot of complexity in the code it avoided!
Multiple fitting schemes
Summary of discussion thus far:
- Existing PIKA code acts on multiple datasets (open/closed/diff), with a single set of parameters in any given Igor experiment:
- Time-series values for
- single ion values
- m/z calibration function and parameters
- instrumental transfer function (PW-PS)
- Single set of values for:
- baseline function parameters (all options in bsl panel) (note this is a time-series in TW)
- set of HR ions that were fit
- set of HR ions that were constrained
- checkbox settings from HR_PeakHeights_Gr panel
- Time-series values for
- PIKA thus will:
- return ONE set of HR sticks PER dataset
- require a re-calculation of HR_Sticks if the parameters are changed
MJC contends that the following are limitations in the current system:
- No data is saved on what parameters led to your currently saved data (a parameter-profile)
- Change a parameter, and the saved data is unaffected
- e.g. one can create baseline-subtracted MS, and then change the m/z cal and carry-on...
- Inability to compare fits calculated using different parameters
- Note that the clunky HR-Sensitivity code simply duplicates data folders to get around this, and relies on a successful execution of the code, as it changes parameters along the way.
DTS important point: However, if one wanted to really allow a user to re-generate the HR_Sticks from the raw spectra, a parameter-profile would also need to save all the steps that went into generating the peak width and peak shape which would be REALLY tedious.
thus
MJC accepts that
- It is not realistic to store a (huge) parameter-profile for a given set of HR_Sticks that would allow the user to reset their experiment to the environment that calculated them.
MJC and DTS disagree on the concept of
- Storing multiple copies of HR-Sticks from any given dataset, calculated using different parameter-profiles.
MJC proposes another compromise solution, that
- The single-MS stage (HR_PeakHeights_Gr) of HR-analysis is totally separated from the time-series calculations,
- At the single-MS stage, users can change the parameter-profile and optimise the HR_Sticks (as currently), but that only a single set of sticks are saved in the time-series,
- The parameter-profiles, including date-time, can be "saved" in some manner during single-MS analysis, to aid in the optimisation,
- During single-MS analysis, we let the user store multiple sets of HR_Sticks for direct intercomparison,
- At the point of calculating time-series, the (advanced) option is presented to save the parameter-profile information in the HDF, and a method to display this is encoded,
- The user is allowed to change the name of the HR_Sticks matrix in the HDF (we need a get-around for open/closed/diff/TWappl etc).
Ion 'bit' indexing
Summary of discussion so far:
- MJC had proposed a bit-indexing scheme that would allow essentially infinite combinations of ion-master-lists to be saved in the fits
- This option is not going to be required, since
- Only one set of time-series sticks will be allowed
- The current master-list system parses a given ion-list at run time and creates an appropriate mask after matching up all the existing selections to the new list (right, DOnna?!)
- The development of a tool to allow comparison of multiple sets of fits using different parameter-profiles would inherently need to store the fitted ion list anyway
MJC thus proposes the solution that
- The existing code to generate mask waves from any given master list is retained
- The existing code to deal with looking for duplicate ions is retained
- The output from the fitting procedure is NOT tied to a master ion list, but an exact mass wave and text wave that corresponds to the ions used in the fit, for
- this is more explicit
- isotopic constraints force the calculation of abundances for every ion in the list, so fitting a subset (and thus wishing to insert/delete points from the base wave) is not an option
- it saves valuable memory for the multi-dimensional datasets
Multiple Dimensions
DTS: The need to add/average PToF raw spectra before HR fits. For AMS PToF data, the signal is so low that I anticipate a grouping/averaging (the AMS size and/or time dimension) is needed of raw spectra before HR fits are attempted. This presents it’s own indexing challenge. For AMS PToF data, one can imagine wanting to average the raw spectra for bins representing 30-100, 100-300, 300-1000nm, for one run before performing the HR fits. (In general it is better to examine the HR fits of an PToF bin-averaged spectra than average the HR fits of noisy raw PToF spectra.) I imagine a similar scenario is true for other Tofwerk ms applications.
MJC: Unfortunately, there are TW applications which will require fitting in two more dimensions than the current AMS-runs... some will want averaging, some will not. As per the m/z calibration, a mask system is going to be required to build up the averaging domains. This is not actually that heinous to put into place and, if we use a MSConcs-esque approach (or even MSConcs itself), the user could specify ANY dimensional base for averaging. The example above would be easily incorporated. But in summary, 3-D fitting is going to be a pre-requisite, along with all the hideous user interface issues this will present afterwards. But the AMS panel need not worry about that! But PTOF data will be able to be fit with the TW scheme...
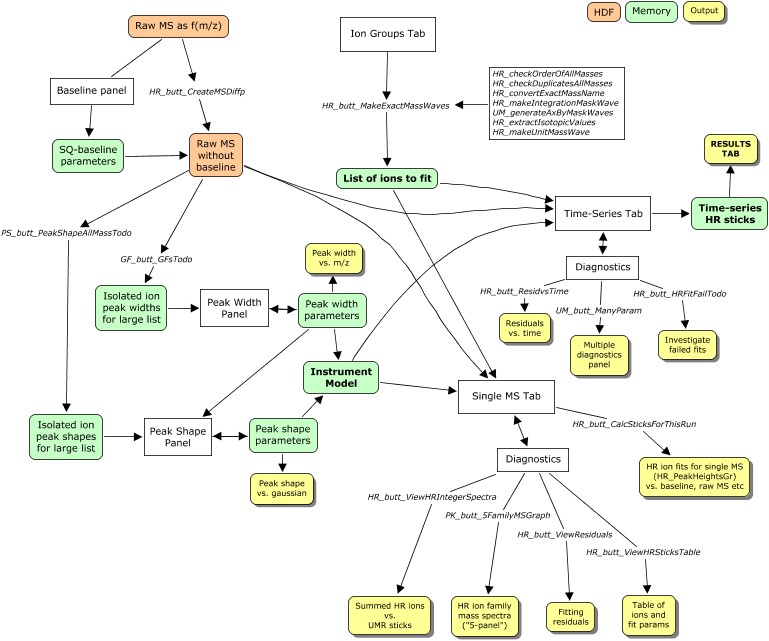
MJC: THIS IS THE POINT OF IMMEDIATE CONCERN. The first step in the PIKA-process (see map below) is to generate the raw MS less baseline... at this point, for multi-dimensional data, the integration and axis-bases (eg t_series for 1-D AMS data) need to be considered. I know JDA has always contended not to integrate mass spectra before fitting, but is this realistic in a low S:N environment? It is clear that MSConcs can generate integrated raw MS (and the baselines) very easily, and I would like to take advantage of this capability. BUT! MSConcs also pulls everything into memory at once... it might be that the solution here is to append to Concs such that it has the capability to write to HDF. Looking at the code, this does not seem very hard. James, what do u think?
DTS: Proposed step forward
A small step forward: ?? Perhaps we can tweak the existing HR code with a modified version of your proposal. Suppose the user really want to get diagnostic information for all runs about whether to fit CHNO;C3H5O;C3H7; or to leave CHNO out. (leaving aside for the moment any isotope constraining issues). Perhaps the code could generate two data sets: HRSticks43_7 and HRSticks43_6. In the AMS world, both data sets would contain at least 3 columns. Version “7” would contain 3 nonzero columns and version “6” 2 nonzero columns. We could id the columns using the HR ion bit-wise idea you have so that column 0 is always CHNO;column 1 is always C3H5O, etc. So the ‘7’ and ‘6’ suffix in the data set name identifies the HR ions fit. This way, a user could examine these multiple versions of HR ions fit at 43.
PIKA fitting process: steps
1. Preparation for HR fitting
The order of preparatory steps below is generally not modifiable. (MJC: comments added for changes required in the TW version)
(1A) Get good m/z calibration parameters. Mike has this code in place for Tofwerk files.
(1B) Get good baseline-removed spectra.
The purpose of saving copies of the raw spectra with the baseline removed is so that the multipeak fitting is done on the ‘same’ spectra. This insures that the calculation of any baseline isn’t dependent on settings (‘resolution’ interpolations parameters) that could be adjusted by the user and then not saved, not
recorded, an hence not replicable.
MJC: Something to consider: should the dimensional averaging be performed at this stage? This would limit the user to the prescribed dimensional bases, but facilitate the sped-up analysis currently available in PIKA.??
DTS: Yes, you are correct. This is the place to generate the spectra that will be fit. I think it important that the PW and PS calcs are based on the same spectra that will be fit.
(1C) Get good peak width (PW), peak shape (PS).
Getting good values for these parameters is necessarily an iterative process. In general one looks at 100s of sets of isolated HR ions (i.e. C4H9+, etc ) spectra to get good statistics for PW and PS. Once a user is confident that selected HR ions are behaving in a consistent and ‘smooth’ manner, one can set the PW and even PS on an individual run basis if needed.
MJC: TW product will require a more generalised version of the current code, which hard-wires in AMS-specific ions...
DTS: Yes the current list of HR ions fit with gaussians is pre-determined, but currently users can add to the list. An import/export list of HR ions to be fit with gaussians is necessary. While on the topic, Jose had always urged that the various lists of ions be linked/merged somehow. That is, from the main list of all ions, there would be flags for each HR ion indicating subsets used for m/z calibration, peak width, peak shape.
(1D) Select the HR ions to fit.
This is highly variable depending on the application. Similar to Mike’s m/z calibration routine, I envision a simple interface whereby a user imports settings appropriate to their type of application.
MJC: Since the UM now I'm wondering about de-constraining the list, too.... hmmm
DTS: I'm not clear what deconstraining the list means. I envision a single prompt to the user at the start of the experiment that would load in their HR ion settings (with flags for m/z calibrations, gaussian fits, peak width, peak shape).
(1E) Select a subset of HR ions in (1D) to be constrained.
Constrained means that the fitting routine does not ‘fit’ this HR ion – the peak height is fixed to a value based on the magnitude of the HR ion’s isotopic ‘parent’ that has been previously fit or determined. This has consequences about the order in which UMR sets of HR ions at on m/z are fit.
2. Perform HR fitting
In the current AMS HR code the HR fitting is performed at one UMR set of HR ions (all chosen HR ions at an individual integer m/z range). For example at nominal m/z 18 the HR ions 18O+, H2O+, 15NH3 at 17.999161, 18.010559, 18.023581 are fit in one ‘set’. Typically isotopic HR ions are constrained, and hence in this case the magnitude of the HR ions of O+ and NH3+ would need to be determined from sets of HR ions at nominal m/z at 16 and 17 before the set of HR ions at 18 would be found. In future applications ToF mass spec applications of high organic fragments (>250m/zish?) with a positive mass defect the division into nominal m/z sets of HR ions could be problematic.
(2A) Single MS high-resolution stick calculation & diagnostics
Before a user spends a lot of computation time generating HR sticks, it is beneficial to examine HR fits of few raw spectra at periods of various concentrations and compositions. This involves visually inspecting each UMR m/z region of interest and significant signal. This visual inspection gives a user feedback on all the parameters that go into the fit: m/z calibration, baseline settings, PW, PS, selection of HR ions to fit. It is often at this stage where at least one of the input parameters of the HR fit requires fine-tuning. Other diagnostics for single spectra include:
- Comparison of HR summed to UMR vs. UMR sticks.
- Residuals from the HR fits.
- “5-panel” graph... Some sort of UMR, family summarized spectra plots. This is more important for EI than for soft or other ionization techniques.
- Tabulated results. Having easy access to the HR sticks in a table is useful for those wishing to check the math or perform their own subsequent calculations.
- Mike’s HR ion sensitivity tool.
(2B) High-resolution stick calculation for many spectra
As the AMS HR code currently exists in version 1.09, the HR fitting results are only saved in intermediate files for future access via subsequent ‘fetch’ commands. It is beneficial for users to have a place to ‘play’ while keeping the HR fitting results from being modified.
3. Organizing, displaying HR stick results & diagnostics
(3A) Organization (order of steps important here)
(3Ai) Define HR families
It will always be convenient to group HR ions into families. A family can be defined explicitly by listing its members (i.e. family Cx =C+, C2+, C3+, etc) or by an algorithm that parses the chemical formula of each HR ion. The AMS HR code currently wants families to be determined at the same time as the selection of the HR ions to fit. However, this is unnecessary and future versions will allow more flexibility.
(3Aii) Define HR batch entities
In AMS parlance a batch entity is typically a species, like organics, nitrate, etc. While the grouping of HR ions into families is convenient, it will always be the case that the chemical information a user desires may require a mathematical or specialized treatment of the HR fit results beyond the simply family sorting mechanism. A common AMS example is the parsing of the OH+ signal between the water and the organic species. Every HR species is defined first by two items: (a) a list of families and (b) a frag wave which identifies any modifications based on individual HR ions (such as the OH example above).
(3Aiii) Define HR frag entries
HR frag entries explicitly states the mathematical treatment of any/all specialized considerations of HR ion, whether the HR ion was fit or not.
(3B) Display
Users will require typical output: time series, mass spectra summed or averaged in user-defined ways, and plotted in a variety of formats. What is different from a UMR analysis is that user have an additional type of entity: HR families as well as individual HR ions, unit resolution summed values, and HR species.
(3C) Diagnostics
All the diagnostics outlined in 2A above, and with a time (or other) dimension will be required by users.