High Resolution ToF-AMS Analysis Guide
Introduction
The High Resolution ToF-AMS (HR-ToF-AMS) Analysis Guide is dedicated to the analysis of complex HR-ToF-AMS field data sets and is intended to establish standard practices in this area on a community-wide basis. It is assumed that users will have some familiarity with Igor and the community based ToF-AMS (Squirrel) analysis tool, Squirrel. Preliminary HR-ToF-AMS data can be generated while a project is underway without worrying about some details; this guide is intended for identifying all post-project analysis steps involved in generating final data.
A Message for Contributors
We want to encourage active participation by all ToF-AMS users in the evolution of the information contained within this wiki and welcome the addition of content that is beneficial to the community as a whole. However, please DO NOT delete any content from this page!! Significant time, effort, and deliberation has gone into the information contained in this page. Rather than deleting content, please feel free to voice your concerns by posting a comment to the discussion page where others can contribute (please be sure to include a topic to be referenced in responses).
HR-ToF-AMS Process Flowchart
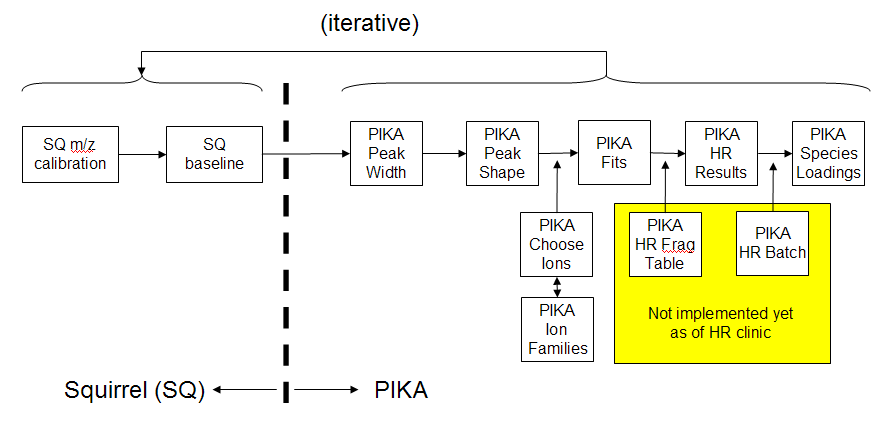
m/z Calibration
- ions have to be isolated at the FWHM level, but not neccesarily at the tail level
- Doug: O+ is often bad, and we don't care about it. We do care about NH2+.
Squirrel Baseline
- Choosing baseline regions
- Check several m/z at low (m/z 15), mid (m/z 50) and high (as high as you care), and see that the baseline regions are proper for all of these regions
- Check for runs at start, middle, and end of experiment (or before and after instrument changes, etc.)
Peak Width
- m/z's used here should be a subset of those used for m/z calibration.
Peak Shape
- ions have to be isolated at the tail level
Choose Ions
- Better to start simple, with fewer ions, and add ions as needed. Having too many ions in the fit can make the fit look good and have low residual for the wrong reasons
- Most people think that the CNO family of ions should NOT be used by default, and only added if really needed per users judgment.
- Web site for exact mass and isotopic calculations
- DeLaeter 2003 IUPAC Atomic Weights paper
- Curators of PIKA Ion List: Puneet Chabra (Chair), Delphine Farmer, Niall Robinson, Qi Chen
Group Ions into Families
- Users can define families for their own purposes
- Each ion can only be a member of one family
PIKA Fits
- Evaluating the fits
- Look at the left side of m/z 48 and 64, which should not have any real ion to their left
- Look at the right side of m/z 41 (C3H5+), which should not have any ions to their right
HR Results
- User should inspect the spectra of each family. Ions that greatly stick out are suspicious, and the user should go back to the PIKA fits and see whether they are real. This is particularly important for the CHN and CHNO ions, which tend to have a very small fraction of the UMR signal at most m/z's.
HR Batch & Frag Tables
This is not implemented yet, but we are targeting to release a version with these components at the 2009 AMS Users Meeting in Toronto. These tables will define the RIE and CE and the fragments to be used to calculate mass concentrations of species from HR data. The frag table is needed to e.g. apportion the H2O+ signal between sulfate, organics, air, and particle water, or the CO2+ signal between air and organics, etc. Right now the user has to do that manually.
PIKA Species Loadings
For the time being, the user has to do that by themselves.